SysBio:
In silico modeling and network construction of chromosomal replication
and segregation
▒ 개 요▒
인간의 염색체복제
및 분리 조절 in silico 시스템 구축 및 제어기술 개발
- 모델 생명체인
효모 및 대장균의 염색체복제 및 분리 네트워크 구성
- 인간
염색체복제 및 분리의 네트워크 구성과 in silico 모델링
- 인위적
조절과 신약개발을 위한 in silico적 응용기술 개발
본 과제는 서울대학교 생명과학부, 생물교육과,
물리학부, 및 연세대학교 생화학과의 공동연구로 이루어지고 있으며 본 팀은
서울대 물리학부와 함께 in silico 모델링을 담당하고 있다.
▒ 제
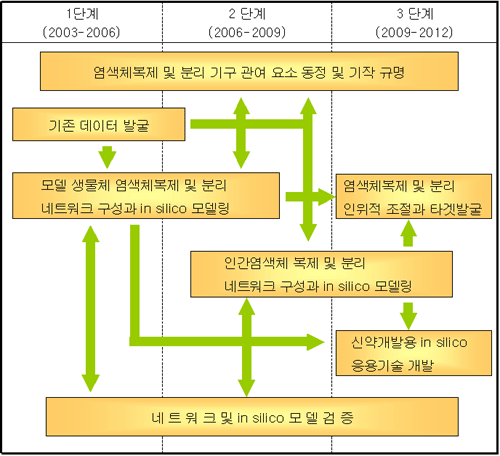
1단계 (2003-2006) 목표 ▒
- 그룹간의
원활한 의사소통, 상호협력 및 학문적 장벽 제거
- 효모
및 대장균의 네트워크 구성과 in silico 모델링
- 염색체복제
및 분리에 관여하는 단백질과 유전자군 발굴 및 기능 규명
- 세포주기
및 유전체 안정성 조절에 관여하는 단백질과 유전자군의 발굴
- 1차년도(2003-2004) -
- Public data를 이용한 생물학적 네트워크의 추출 및 특성 분석/이해
- Cell-cycle 관련 유전자들의 prototype 네트워크 구축
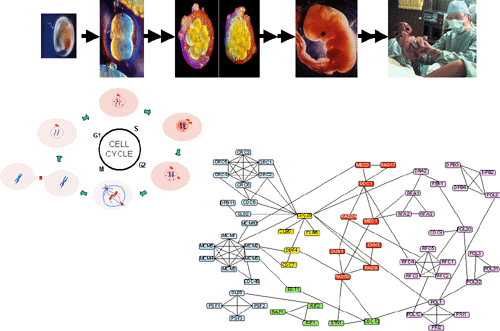
연구성과: MIPS의 PPI 네트워크 구조
노드-링크 개수간의 멱급수 분포 구조 및 그래프 이론에 근거하여 Hub node (중요 유전자)발견 및 Funtional grouping을 시도, 네트워크를 분석/이해함
- 2차년도(2004-2005) -
- Celluar dynamics를 나타내는 데이터를 표현할 수 있는 유전자 네트워크의 구성
- Cell-cycle 유전자 네트워크를 전체 네트워크 측면에서 분석, 중요 유전자군 추출
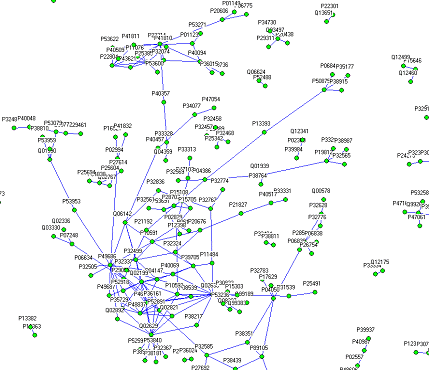
연구성과: MRE를 이용한 S.Cerevisiae의 time-series, PPI 혼합 네트워크 구축
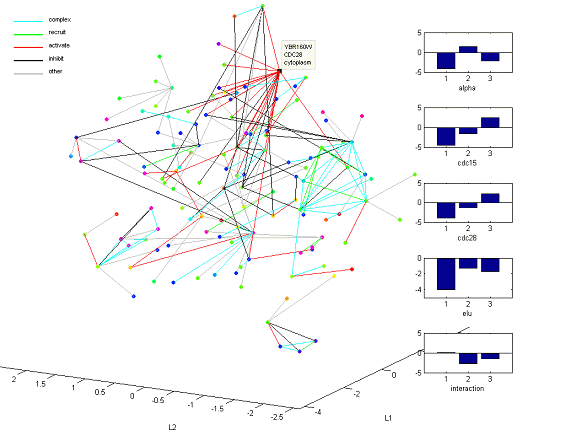
Celluar Dynamic를 표현하는 데이터의 일종인 Microarray time-series 데이터와 PPI (protein-protein interaction) 테이터를 결합하여 네트워크를 구축할 수 있는 방법론을 개발함
▒ 연구
추진 계획 ▒
|
▒ Publications ▒
- 1단계 2차년도(2004.6~2005.3) -
- Searching transcriptional modules using evolutionary algorithms, J.-G. Joung,
S. J. Oh, B.-T. Zhang, Lecture
Notes in Computer Science, 3242, 532-540, 2004
- Computational methods for identification of human microRNA precursors, J.-W. Nam, W.-J. Lee, B.-T. Zhang, Lecture
Notes in Artificial Intelligence, 3157, 732-741, 2004
- Prediction of implicit protein-protein interaction by optimal associative feature mining, J.-H. Eom,
J.-H. Chang, B.-T. Zhang, Lecture
Notes in Computer Science, 3177, 85-91, 2004
- Prediction of the Risk Types of Human Papillomaviruses by Support Vector Machines, J.-G. Joung, S. J. O, and B.-T. Zhang, Lecture Notes in Artificial Intelligence, 3157:723-731, 2004.
- Protein Sequence-based Risk Classification for Human Papillomaviruses, J.-G. Joung, S. J. O and B.-T. Zhang, Computers in Biology and Medicine, (to appear).?
- PubMiner: Machine learning-based text mining system for biomedical information mining, J.-H. Eom and B.-T. Zhang, Lecture Notes in Artificial Intelligence, 3192:216-225, 2004.
- Genetic Mining of DNA Sequence Structures for Effective Classification of the Risk Types of Human Papillomavirus(HPV), J.-H. Eom, S.-B. Park, and B.-T. Zhang, Lecture Notes in Computer Science, 3316:1334-1343, 2004.
- BioPubMiner: Machine Learning Component-based Biomedical Information Analysis Platform, Eom, J.-H. and Zhang, B.-T., Lecture Notes in Computer Science, vol. 3356, pp. 11-20, 2004.
- Adaptive Neural Network based Clustering of Yeast Protein-Protein Interactions, Eom, J.-H. and Zhang, B.-T., Lecture Notes in Computer Science, vol. 3356, pp. 49-57, 2004.
- 1단계 1차년도(2003.7~2004.6) -
- 정보이론에
기반한 Supervised, Unsupervised 피처 선택 방법론,
이상근, 김병희, 장병탁, 한국정보과학회 제31회
춘계학술대회, 2004.
- 진화
알고리즘을 통한 전사 조절 모티프 조합 탐색,
이제근, 정제균, 장병탁, 한국정보과학회 제31회
춘계학술대회, 2004.
- Two-step
Genetic Programming for Optimization of RNA
Common-structure, Jin-Wu Nam, Je-Gun Joung,
Byoung-Tak Zhang, EVOBIO, 2004.
- 인공신경망을 이용한 세포 주기상의 전사 조절 모티프 탐색, 이제근, 정제균, 장병탁, 한국정보과학회 제31회 추계학술대회, 2004
- 재구성된
유전자 네트워크의 섭동적(perturbational) 토폴로지
변형 분석, 이상근, 장병탁, 한국정보과학회
제30회 추계학술대회, 30(2), 2003.
- 생화학
시스템의 동적 모델링을 위한 S-tree 기반의 진화연산,
조동연, 장병탁, 한국정보과학회 제30회 추계학술대회, 30(2),
2003.
|
|
Project Title |
In silico modeling and network construction
of chromosomal replication and segregation
|
|
Sponsor |
The Ministry of Science and
Technology of Korea |
|
Principal Investigator |
Prof.Byoung-Tak Zhang
|
|
Researchers |
Je-Gun Joung
Jin-Wu Nam
Sang Kyun Lee
Je-Keun Rhee
Sung-Kyu Kim
|
|
Duration |
2003 - 2012 |
|
Cooperative Research Institutes |
School of Biological Sciences,
School of Physics, and Department of Biology Education,
Seoul National University;
School of Biochemistry, Yonsei
University |
|
|



